Step 2. Then, we start refining the cladogram by progressively adding more characters to it. The choice of the first character to start with is somehow subjective, but as a general rule you should always start with binary characters (those with only 0 or 1 as a value) and characters that appear derived in a large number of species (coded ”1” in the matrix). Such characters define large monophyletic groups.
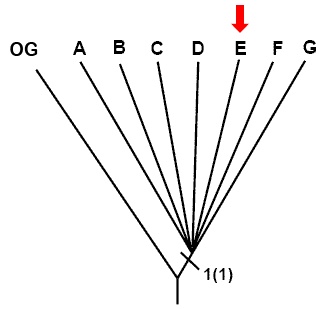
In our example, character #1 indicates that all fish species of the ingroup have an operculum (bone that covers the branchial chamber) while the outgroup possesses slits on the sides of the branchial cavity. Therefore, the no resolution cladogram can be modified to reflect the distribution of character 1 states, and therefore the position of the transformation event from state 0 to state 1.
To do so, a new branch is created, separating the OG that possesses the state (0), and taxa [A-G] that possess the derived state (1). The transformation of character 1 is represented with a tick mark on the new branch, which is labelled with the character number followed by the new state of this character between brackets: 1(1). This creates a new monophyletic group: [A-G].
Cladogram 2